Plot Character History
plot.history.RdBoth stochastic character mapping and simulation may create character histories. This function plots these histories
Arguments
- x
An object of class
history.discretecontaining a discrete character history. This could be made byhistory.from.sim.discrete.- phy
The phylogeny used to generate the history. Few checks are made to make sure that this is really correct, and all manner of terrible things might happen if these are not compatible. This may change in future.
- cols
A vector of colours.
- states
The different state types. Probably best to leave alone.
- xlim
Plot x-limits (optional).
- ylim
Plot y-limits (optional).
- show.tip.label
Logical: show the species tip labels?
- show.node.label
Logical: show the species node labels?
- show.tip.state
Logical: draw a symbol at the tips to indicate tip state?
- show.node.state
Logical: draw a symbol at the nodes to indicate node state?
- no.margin
Supress drawing of margins around the plot
- cex
Font and symbol scaling factor.
- font
Font used for tip and node labels (see
par).- srt
String rotation for tip and node labels.
- adj
Label
adjustment (seepar).- label.offset
Horizontal offset of tip and node labels, in branch length units.
- lwd
Line width
- ...
Additional arguments (currently ignored)
Details
This attempts to be as compatible with ape's plotting functions
as possible, but currently implements only right-facing cladegrams.
Examples
## Due to a change in sample() behaviour in newer R it is necessary to
## use an older algorithm to replicate the previous examples
if (getRversion() >= "3.6.0") {
RNGkind(sample.kind = "Rounding")
}
#> Warning: non-uniform 'Rounding' sampler used
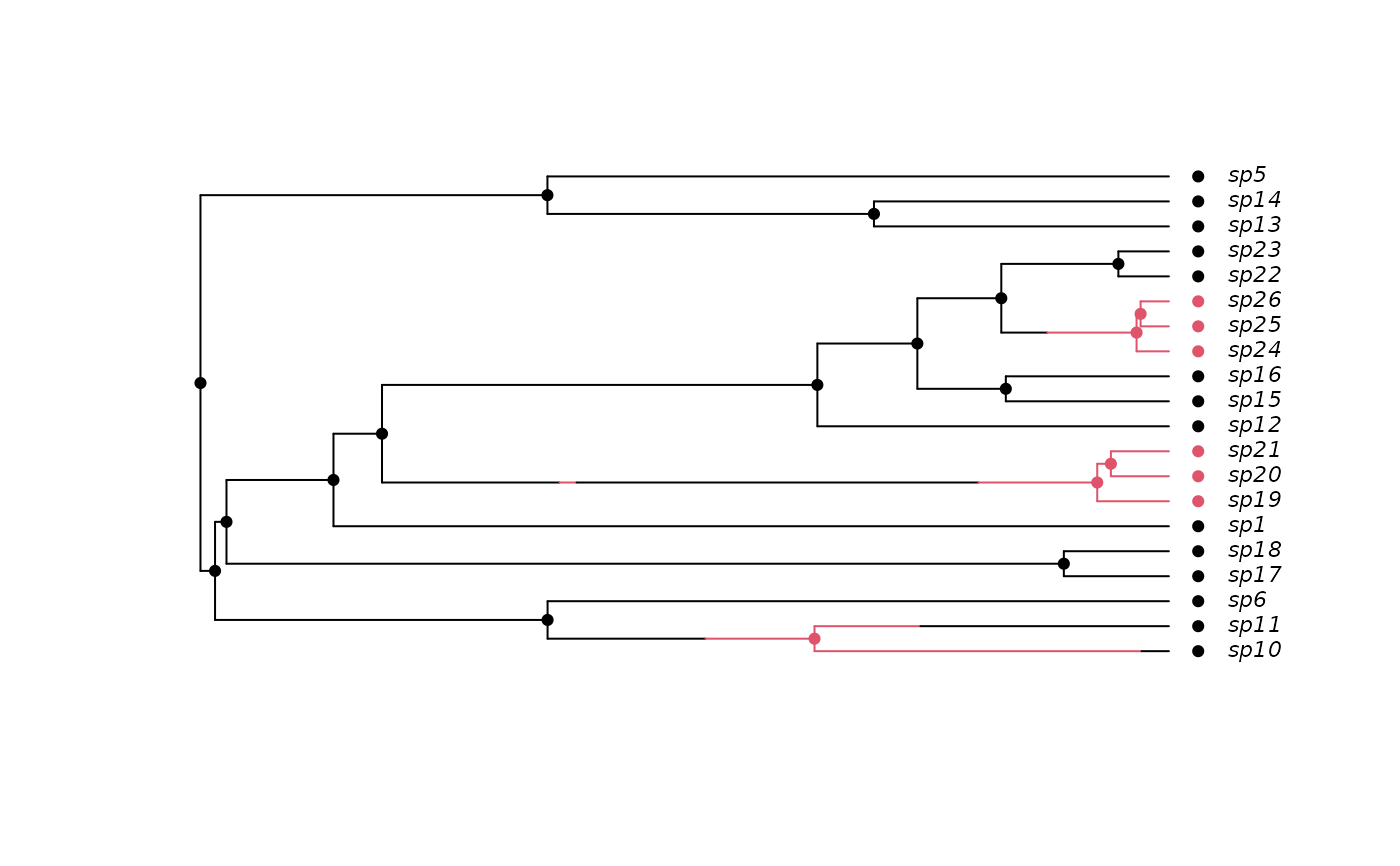
## Simulate a tree, but retain extinct species.
pars <- c(.1, .2, .03, .04, 0.05, 0.1) # BiSSE pars
set.seed(2)
phy <- tree.bisse(pars, 20, x0=0, include.extinct=TRUE)
## Create a 'history' from the information produced by the simulation
## and plot this
h <- history.from.sim.discrete(phy, 0:1)
plot(h, phy, cex=.7)
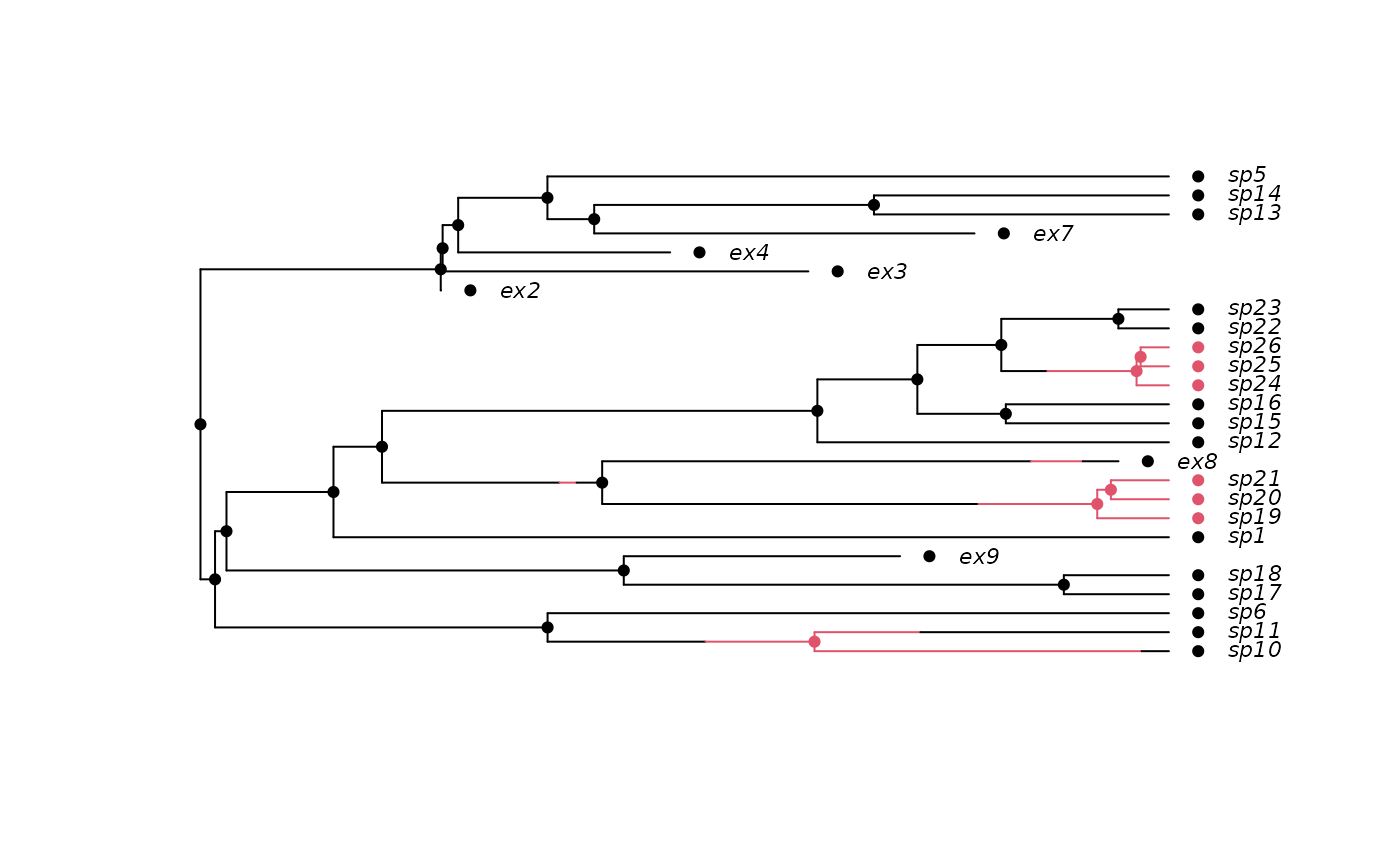 ## Prune the extinct taxa.
phy2 <- prune(phy)
## The history must be recreated for this pruned tree:
h2 <- history.from.sim.discrete(phy2, 0:1)
plot(h2, phy2, cex=.7)
## Prune the extinct taxa.
phy2 <- prune(phy)
## The history must be recreated for this pruned tree:
h2 <- history.from.sim.discrete(phy2, 0:1)
plot(h2, phy2, cex=.7)